News & press releases
LEGaTO research on hardware accelerators for DNA-comparisons
While LEGaTO mainly focuses on its main use-cases like Smart Home, Smart City or Machine Learning, other applications are also analyzed concerning their performance and energy efficiency, in particular FPGA-centric applications. In cooperation with the Queensland University of Technology (Australia), the research team from Bielefeld University analyzed the acceleration of binary string comparisons on FPGAs using a new approach for a scalable, streaming-based system architecture.
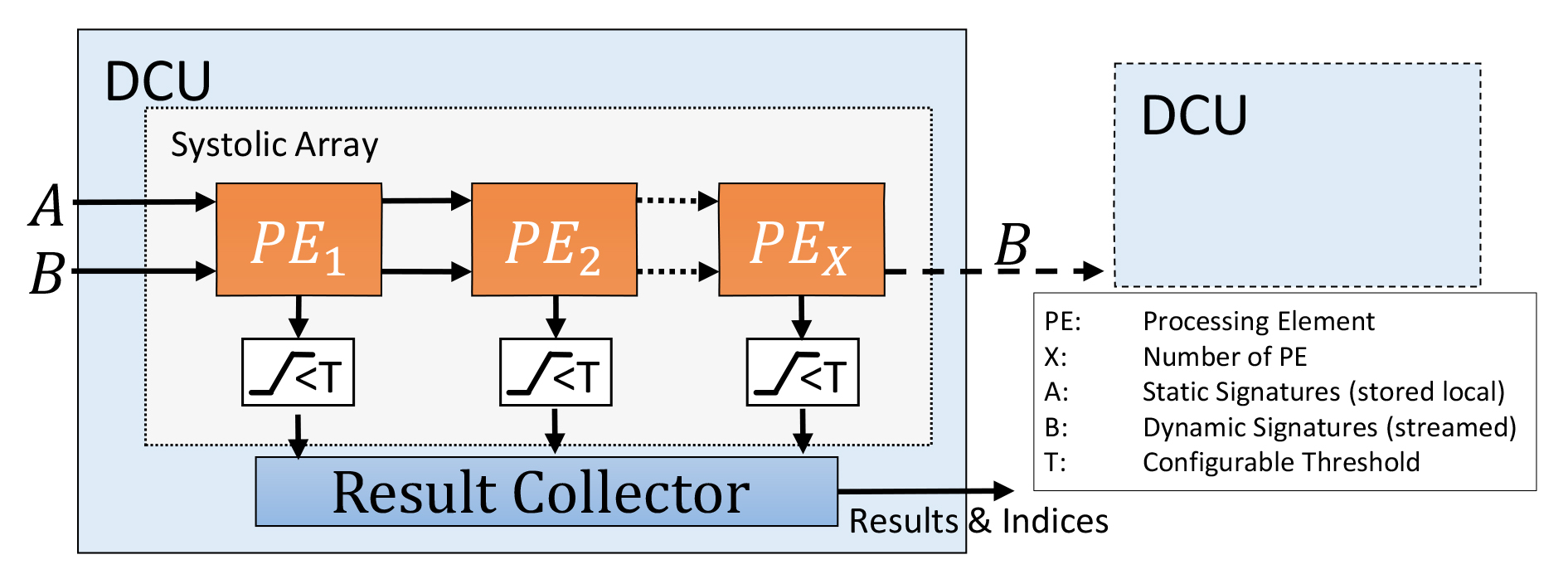
The primary focus of the work is to accelerate the calculation of hamming distances, which is the primary source of the load for Locality Sensitive Hashing. This algorithm can be used to compare, e.g., the similarity of DNA sequences. In this application, typically hundreds of queries are compared with large data sets, resulting in billions of calculations.
Although the algorithm for Hamming Distance comparison is rather simple, achieving a highly parallel and scalable implementation was quite a challenging task. The design, described in VHDL, was executed using a Xilinx VCU1525 evaluation board featuring an UltraScale+ VU9P FPGA. A peak throughput of 75.4 billion comparisons per second - of 512-bit signatures - was achieved, using a design with 384 parallel processing elements and a clock frequency of 200 MHz. This makes the proposed FPGA design 86 times faster than a highly optimized CPU implementation. Compared to a GPU design, executed on an NVIDIA GTX1060, it performs nearly five times faster.
Based on this preliminary work, we plan to port the hardware accelerator to the upcoming Edge Server t.RECS developed within LEGaTO, which will support FPGA modules with UltraScale+ SoC devices. For future work, the design, which was created via low-level tools, could be re-implemented using the high-level LEGaTO stack using the OmpSs programming model or the DFiant HDL approach.
The full text of the related publication is open access and can be accessed using the link below:

